Research projects
- On this page:
- Selected project narratives
- Other project summaries:
- 3D microscopy
- Single-molecule orientation
- Microscope on a chip
View my lab's current research projects on the Lew Lab website.
Selected project narratives
Simultaneously measuring the 3D position and orientation of single molecules

We study the complex radiation pattern of single molecules (modeled as electric dipoles), which is often neglected in traditional single molecule-based super-resolution experiments. Ignoring this radiation pattern can produce sizable errors in measurements of single-molecule position, but using a behavior uniquely inherent in the double-lobed nature of the double-helix point spread function, we account for such mislocalizations and simultaneously measure 3D molecular orientation and 3D position. Read or watch project summary video
M. P. Backlund*, M. D. Lew*, A. S. Backer, S. J. Sahl, G. Grover, A. Agrawal, R. Piestun, and W. E. Moerner, "Simultaneous, accurate measurement of the 3D position and orientation of single molecules," Proc. Natl. Acad. Sci. USA 109, 19087 (2012). [Highlight in Nat. Methods, Journal, PDF]
Imaging the bacterial cell surface and an internal protein in 3D
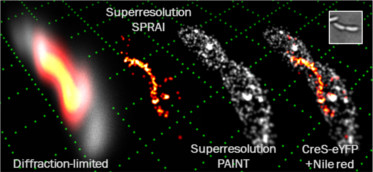
SPRAIPAINT is a technique that combines imaging of an internal fluorescent protein (SPRAI) with random sampling of the cell surface (PAINT, developed by Hochstrasser et al.). The SPRAIPAINT scheme achieves single-molecule localizations without any antibody labeling, cell membrane permeabilization, or thiol-oxygen scavenger systems. Here we demonstrate 3D super-resolution imaging of a cytoskeletal protein colocalized with the surface of a living bacterium using the double-helix microscope. Read or watch project summary video
M. D. Lew*, S. F. Lee*, J. L. Ptacin, M. K. Lee, R. J. Twieg, L. Shapiro, and W. E. Moerner, "Three-dimensional superresolution colocalization of intracellular protein superstructures and the cell surface in live Caulobacter crescentus," Proc. Natl. Acad. Sci. USA 108, E1102 (2011). [Journal, PDF]
3D super-resolution microscopy
Bending light into a corkscrew shape for 3D imaging of point-like objects
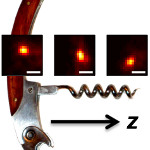
We redesign a microscope's imaging behavior so that instead of an object simply being either in focus (sharp) or out of focus (blurry), it is in focus over a large range of depths. Within this depth range, the image of the object revolves around a central axis. If the object is moved away or toward the lens, then the image of the object revolves to trace out a corkscrew shape, hence the name of the microscope: the corkscrew microscope. By precisely measuring the position and rotation of the image of any point-like object, we can easily measure its 3D position.
M. D. Lew, S. F. Lee, M. Badieirostami, and W. E. Moerner, "Corkscrew point spread function for far-field three-dimensional nanoscale localization of pointlike objects," Opt. Lett. 36, 202 (2011). [Journal, PDF]
The double-helix microscope: 3D precision measurements and its tracking applications
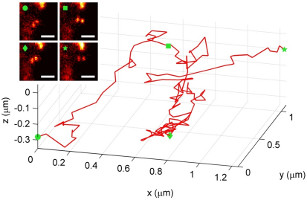
Here, we demonstrate measuring and tracking the 3D position of single dim objects with a double-helix fluorescence microscope. This microscope gets its name from the bending of the light emitted from each point-like object into a double-helix shape; as an emitter moves closer or further away from a lens, the image of the emitter forms two spots that revolve around one another on a camera, tracing out a double-helix along the third dimension. We describe in detail how the precision of the microscope quantitatively depends upon the number of photons detected and the depth of the nanoscale emitter. The double-helix microscope, for the first time, is used to track a quantum dot-labeled structure inside a living cell in three dimensions.
M. A. Thompson*, M. D. Lew*, M. Badieirostami, and W. E. Moerner, "Localizing and tracking single nanoscale emitters in three dimensions with high spatiotemporal resolution using a double-helix point spread function," Nano Lett. 10, 211 (2010). [Journal]
Single-molecule orientation studies
Rotational wobble of single molecules affects accuracy in super-resolution microscopy
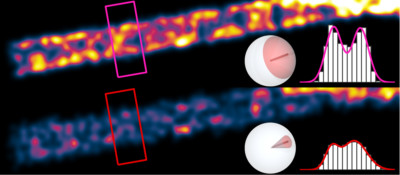
Single molecules resemble oscillating electric dipoles with a toroidal light emission pattern. If this dipole-like behavior is ignored, there can be significant accuracy errors in single molecule-based super-resolution microscopy. The degree of error depends highly on the rotational mobility, or "wobble", of each molecule. Our simulations show that relatively fixed molecules cause distortions in super-resolution microscope images. In certain conditions, they can even degrade the resolution of these images.
M. D. Lew*, M. P. Backlund*, and W. E. Moerner, "Rotational mobility of single molecules affects localization accuracy in super-resolution fluorescence microscopy," Nano Lett. 13, 3967 (2013). [Journal cover, Journal]
Microscope on a chip
Measuring wavefront tilt directly on a camera chip without any lenses
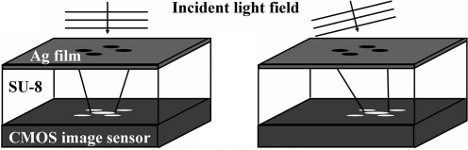
We present a new way to measure the tilt of the wavefronts of a light field using four circular holes arranged in a "plus" pattern above a standard CMOS camera chip. The diffraction pattern of the aperture, as seen by the sensor, moves as the incoming wavefront becomes increasingly inclined. We use this device to measure the wavefront of a Gaussian laser beam and an optical vortex.
M. Lew, X. Cui, X. Heng, and C. Yang, "Interference of a four-hole aperture for on-chip quantitative two-dimensional differential phase imaging," Opt. Lett. 32, 2963 (2007). [Journal, PDF]