Imaging the bacterial cell surface and an internal protein in 3D
M. D. Lew*, S. F. Lee*, J. L. Ptacin, M. K. Lee, R. J. Twieg, L. Shapiro, and W. E. Moerner, "Three-dimensional superresolution colocalization of intracellular protein superstructures and the cell surface in live Caulobacter crescentus," Proc. Natl. Acad. Sci. USA 108, E1102 (2011). [Journal, PDF]
Project summary video
"Three-dimensional super-resolution co-localization of intracellular protein superstructures and the cell membrane in live Caulobacter crescentus," 3rd Annual Center for Biological Imaging at Stanford Symposium, Stanford, CA, March 2011.
Introduction
As much as research and development is driven by long-term planning and forethought, sometimes serendipity is just as important. One of the most important lessons this project has taught me is to seize the opportunities that present themselves entirely by accident.
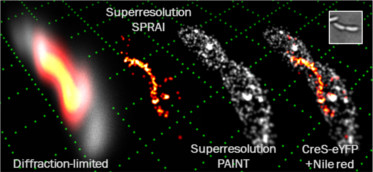
A then postdoc in our lab, Dr. Steven Lee, was imaging some wild-type bacteria in the microscope as a negative control. These cells should have been non-fluorescent since no genetic modifications to express fluorescent proteins had been performed on them. However, he found that they were bright, and when he analyzed the movies he took of the blinking single molecules, he found that, miraculously, the super-resolution reconstructions highlighted the cell surface with high precision! Thus, 3D SPRAIPAINT was born.
Importance
SPRAIPAINT (Superresolution by PoweR-dependent Active Intermittency and Points Accumulation for Imaging in Nanoscale Topography) is the first demonstration of 3D super-resolution imaging of a biological structure colocalized with the surface of a a living bacterial cell. It generates two-color images through a unique two-step process: 1) imaging of a fluorescent protein tagged to a biological structure of interest (in this case, enhanced yellow fluorescent protein [EYFP] labeling the Crescentin cytoskeleton) followed by 2) imaging of an organic fluorescent molecule that blinks on only when near the cell surface (Nile Red). It proves that the double-helix (DH) microscope can be used to generate super-resolution images of structures within living cells despite criticism that the microscope is photon inefficient and that the PSF is to large. This project is also the first time that I used the DH microscope to produce 3D super-resolution images.
Tasks completed
- We optimized the expression levels of the CreS-EYFP fusion to maximize fluorescent signal without making the cells sick.
- We tuned the buffer conditions for Nile Red PAINT to maximize signal to background ratio and label specificity.
- We characterized the power-dependence of EYFP blinking and proposed a simple kinetic model that describes the observed dynamics.
- We wrote automated image processing code to recognize DH images in raw movies and extract 3D positions from each single-molecule blinking event that passes certain filters for image quality. We tested the algorithm against the molecules recognized by a human to ensure accuracy.
- Our article was published in Proc. Natl. Acad. Sci. USA and presented at several conferences.
Skills gained
- Basic kinetic modeling of the electronic transition rates of a fluorescent molecule
- Image processing techniques including filtering, Fourier domain processing, and template matching
- Preparation and immobilization of bacteria on agarose pads for live-cell imaging
- 3D rendering of single-molecule localization data in both still images and animated movies
Lessons learned
- When trying a new experiment, be sure to explore the parameter space widely in order to find the conditions that give the best data quality. We tried different species of bacteria, imaging buffers, fluorophores, pumping intensities, camera exposure times, fluorescence filters, etc. to collect the best possible data.
- Creating and generating the meaning of acronyms are fun! SPRAYPAINT was our original idea (PAINT having been published years before), but we couldn't come up with a suitable word beginning with a "Y".
- With proper tuning, automated image recognition algorithms can perform quite well in the presence of noise. This saves many man-hours over manual human recognition of each single molecule one by one!
- A great project partner makes the difference between a productive, collaborative experience and an unpleasant, conflict-filled trial by fire. Steve and I split the project tasks, taking care of the items we were each best-suited for, and we completed the project without worrying about individual turf and who would get credit. I would work with Steve again in a heartbeat!
Impact
By writing the automated image recognition algorithm, it now no longer takes days to analyze a single-molecule super-resolution dataset, increasing our productivity by an order of magnitude. This software is now available as a free open-source download. The labeling and imaging techniques vetted by this project laid the foundation for future 3D super-resolution imaging of proteins within bacteria by the Moerner lab. The images we obtained during this project solidified the DH microscope in my mind as a reliable, accurate microscope viable for answering fundamental questions in cell biology. With further refinements, it has great potential to become the workhorse microscope in biological imaging laboratories.
Related work:
- M. D. Lew, A. R. S. von Diezmann, and W. E. Moerner, "Easy-DHPSF open-source software for three-dimensional localization of single molecules with precision beyond the optical diffraction limit," Protocol Exchange (2013). [Journal, PDF]
- H-L. D. Lee*, S. J. Sahl*, M. D. Lew, and W. E. Moerner, "The double-helix microscope super-resolves extended biological structures by localizing single blinking molecules in three dimensions with nanoscale precision," Appl. Phys. Lett. 100, 153701 (2012). [Journal cover, Journal]